About Disorder Atlas
Background
Intrinsic disorder is a property describing proteins and protein regions lacking a stable three-dimensional structure under physiological conditions. The experimental challenges imposed by this property has led to the development of large number of computational tools capable of predicting disorder in protein sequences. These algorithms often employ different tactics toward identifying intrinsic disorder, which commonly includes the assessment of physicochemical properties and/or utilizing sequence alignment against a database of disordered proteins. When analyzing the results users are faced with the problem of interpreting the predicted disorder content of a protein. The first step of interpretation often involves calculating relevant descriptive statistics from disorder propensity information, such as percent disorder, as well as the length and location of continuous stretches of disordered residues. However, interpreting the meaning of these statistics for single proteins remains difficult. For example, if ‘protein X’ is predicted to contain a disorder content of 20%, how does one evaluate whether this feature is significant?
Disorder Atlas is a web-based service that facilitates the interpretation of intrinsic disorder predictions using proteome-based descriptive statistics. This service is also equipped to facilitate large-scale systematic exploratory searches for proteins encompassing disorder features of interest, and further allows users to browse the prevalence of multiple disorder features at the proteome level.
Is Disorder Atlas based on past work?
Yes, Disorder Atlas is based on the analysis of Vincent et al., 2016, which sought to provide quantitative guidelines, specific to both proteomes and disorder prediction algorithms, for objectively assessing intrinsic disorder. Using a rigorous non-parametric statistical approach, Vincent et al., 2016 provided standards for the following disorder metrics in ten eukaryotic proteomes:
Disorder content:
The percentage of disordered residues contained in a primary amino acid sequence.
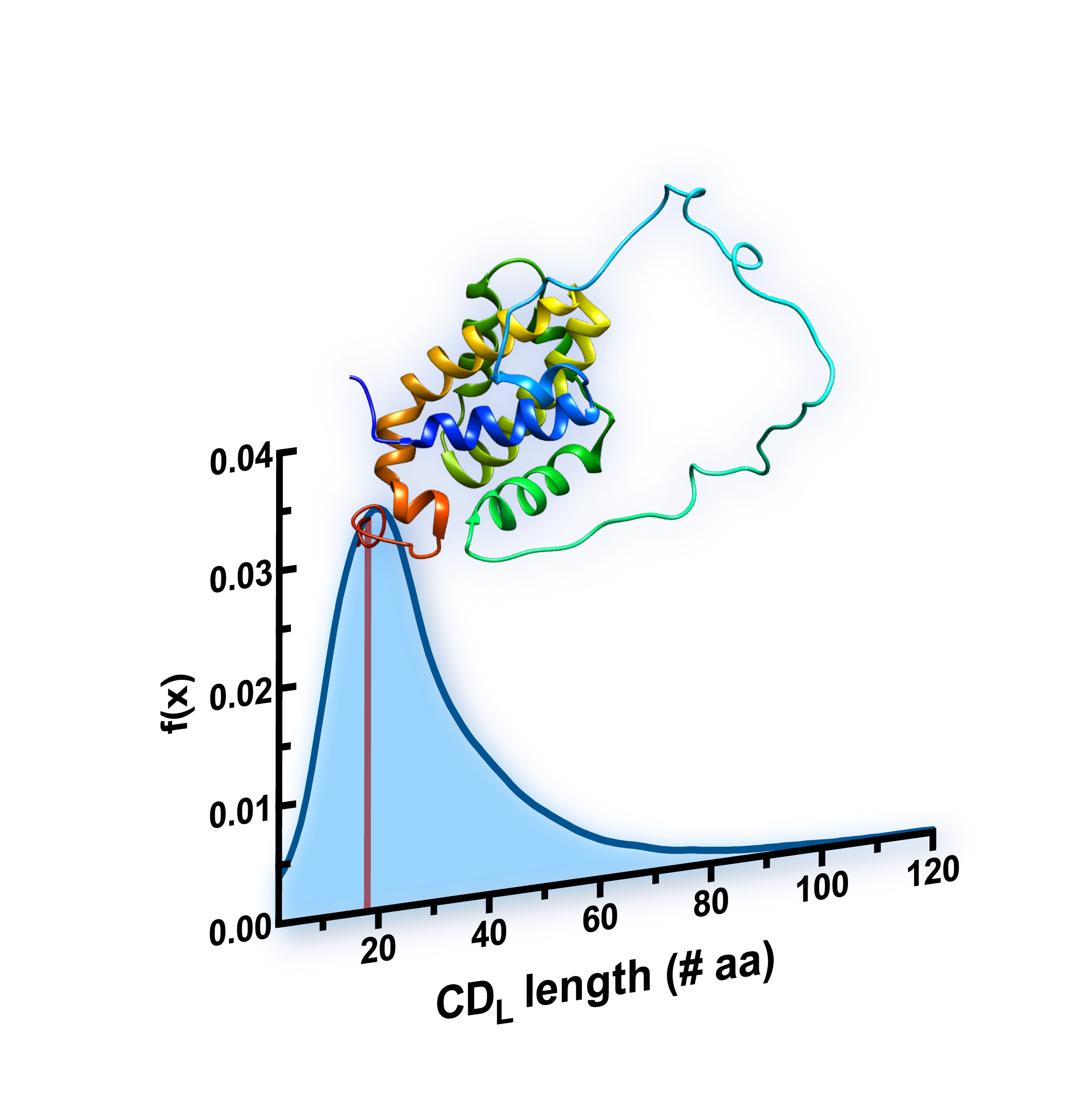
Longest continuous disorder region ( CDL ):
The longest continuous stretch of disordered amino acids found in a protein.
Longest CDL percentage of length ( LCPL ):
The percentage of the protein length accounted for by the CDL.
What algorithms are supported by Disorder Atlas?
Disorder Atlas supports a number of algorithms that predict intrinsic disorder, as well as other parameters relevant to disorder prediction (charge distribution, hydropathy, etc.). A complete list of supported algorithms can be found here.